Using Epigrass¶
To simulate an epidemic process in Epigrass, the user needs to have in hand at least three files: Two files containing the site and edge data and a third file which is a script that defines what it is to be done. Here we go through each one of them in detail. The last part of this chapter is a step-by step guide the Graphical User Interface (GUI).
Data¶
Site data file¶
See below an example of the content of a site file for a network of 10 cities. Each line corresponds to a site (except the first line which is the title). For each site, it is declared, in this order: its spatial location in the form of a pair of coordinates ([X,Y]); a site $name$ to be used in the output; the site’s population; the site geocode (an arbitrary unique number which is used internally by Epigrass).
| X | Y | City | Pop | Geocode |
|---|---|---|---|---|
| 1 | 4 | “N1” | 1000000 | 1 |
| 2 | 4 | “N2” | 100000 | 2 |
| 3 | 4 | “N3” | 1000 | 3 |
| 4 | 4 | “N4” | 1000 | 4 |
| 5 | 4 | “N5” | 1000 | 5 |
| 1 | 3 | “N6” | 100000 | 6 |
| 2 | 3 | “N7” | 1000 | 7 |
| 3 | 3 | “N8” | 100000 | 8 |
| 4 | 3 | “N9” | 100000 | 9 |
| 5 | 3 | “N10” | 1000 | 10 |
| 1 | 2 | “N11” | 1000 | 11 |
In this example, the first site is located at [X,Y]=[1,4], it is named N1, its population is 1000000 and its geocode is 1. This is minimum configuration of a site data file and it must contains this information in exactly this order.
In some situations, the user may want to add other attributes to the sites (different transmission parameters, or vaccine coverage or initial conditions for simulations). This information is provided by adding new columns to the minimum file. For example, if one wishes to add information on the vaccine coverage in cities N1 to N10 ($vac$) as well as information about average temperature (which hypothetically affects the transmission of the disease), the file becomes:
| X | Y | City | Pop | Geocode | Vac | Temp |
|---|---|---|---|---|---|---|
| 1 | 4 | “N1” | 1000000 | 1 | 0.9 | 32 |
| 2 | 4 | “N2” | 100000 | 2 | 0.88 | 29 |
| 3 | 4 | “N3” | 1000 | 3 | 0.7 | 25 |
| 4 | 4 | “N4” | 1000 | 4 | 0.2 | 34 |
| 5 | 4 | “N5” | 1000 | 5 | 0 | 26 |
| 1 | 3 | “N6” | 100000 | 6 | 0 | 27 |
| 2 | 3 | “N7” | 1000 | 7 | 0 | 31 |
| 3 | 3 | “N8” | 100000 | 8 | 0 | 30 |
| 4 | 3 | “N9” | 100000 | 9 | 0 | 24 |
| 5 | 3 | “N10” | 1000 | 10 | 0 | 31 |
During the simulation, each site object receives these informations and store them in appropriate variables that can be used later during model specification. Population is stored in the variable N; while the extra columns (those beyond the geocode) are stored in a tuple named {values. For example, for the city N1, we have N = 1000000 and values=[0.9,32]. During model specification, we may use N to indicate the population size and/or we can use values[0] to indicate the level of vaccination of that city and values[1] to indicate the temperature.
It is up to the user, to know what means the elements of the tuple values. Note that the first element of the tuple has index 0,the second one has index 1 and so on.
When using real data, one may wish to use actual geocodes and coordinates. For example, for a network of Brazilian cities, one may build the following file:
| latitude | longitude | local | pop | geocode |
|---|---|---|---|---|
| -16:19:41 | -48:57:10 | ANAPOLIS | 280164 | 520110805 |
| -10:54:32 | -37:04:03 | ARACAJU | 461534 | 280030805 |
| -21:12:27 | -50:26:24 | ARACATUBA | 164449 | 350280405 |
| -18:38:44 | -48:11:36 | ARAGUARI | 92748 | 310350405 |
| -21:13:17 | -43:46:12 | BARBACENA | 103669 | 310560805 |
| -22:32:53 | -44:10:30 | BARRA_MANSA | 165134 | 330040705 |
| -20:33:11 | -48:34:11 | BARRETOS | 98860 | 350550005 |
| -26:54:55 | -49:04:15 | BLUMENAU | 241943 | 420240405 |
| -22:57:09 | -46:32:30 | B.PAULISTA | 111091 | 350760505 |
In this file, the coordinates are the actual geographical latitude and longitude coordinates. This information is important when using Epigrass with a map in shapefile format. The geocode is also the official geocode of these localities (the same one used in the shapefile). Despite the cumbersome size of the number, it may be worth using it because demographic official databases are often linked by this number.
Edge data file¶
The edge data file contains all the direct links between sites. Each line in the file (except for the first, which is the header) corresponds to an edge. For each edge (or link) one must specify (in this order): the names of the sites connected by that edge; the number of individuals traveling from source to destination; the number of individuals travelling from destination to source per time step; the distance or length of the edge. At last, the file must contain, in the fifth and sixth columns, the geocodes of the source and destination sites*. This is very important as the graph is built internally connecting sites through edges and this is done based on geocode info.
Warning
It is required that the order of columns is kept the same.
See below the list of the 8 edges connecting the sites N1 to N10. Let’s look the first one, as an example. It links N1 to N2. Through this link passes 11 individuals backwards and forwards per time step (a day, for example). This edge has length 1 (remember that N1 is at [X,Y]=[1,1] and N2 is at [X,Y]=[1,2], so the distance between them is 1). The last two columns show the geocode of N1 (geocode 1) and the geocode of N2 (geocode 2).
| Source | Dest | flowSD | flowDS | Distance | geoSource | geoDest |
|---|---|---|---|---|---|---|
| N1 | N2 | 11 | 11 | 1 | 1 | 2 |
| N2 | N4 | 0.02 | 0.02 | 1 | 3 | 4 |
| N3 | N8 | 1.01 | 1.01 | 1 | 3 | 8 |
| N4 | N9 | 1.01 | 1.01 | 1 | 4 | 9 |
| N5 | N10 | 0.02 | 0.02 | 1 | 5 | 10 |
| N6 | N5 | 1.01 | 1.01 | 1 | 7 | 8 |
| N7 | N10 | 1.01 | 1.01 | 1 | 7 | 8 |
| N9 | N10 | 1.01 | 1.01 | 1 | 9 | 10 |
Note that it doesn’t matter which site is considered a Source and which one is considered a Destination. I.e., if there is a link between A and B, one may either named A as source and B as destination, or the other way around.
If the edge represents a road or a river, one may use the actual metric distance as length. If the edge links arbitrary localities, one may opt to use euclidean distance, calculated from the x and y coordinates (using Pythagoras theorem).
Specifying a model: the script¶
Once the user has specified the two data files, the next step is to define the model to be executed. This is done in the .epg script file. The .epg script is a text file and can be edited with any editor (not word processor). This script must be prepared with care.
The best way to write down your own .epg is to edit an already existing .epg file. So, open Epigrass, choose an .epg file and click on the Edit button. Your favorite editor will open and you can start editing. Don’t forget to save it as a new file in your working directory. Of course, there is an infinite number of possibilities regarding the elaboration of the script. It all depends on the goals of the user.
Note
Another way to edit an .epg file is to open it whith the graphical editor provided with Epigrass. Just type epgeditor yourmodel.epg.
For the beginner, we suggest him/her to take a look at the .epg files in the demo directory. They are all commented and may help the user in getting used with Epigrass language and capabilities.
Some hints to be successful when editing your .epg:
- All comments in the script are preceded by the symbol #. These comments may be edited by the user as he/she wishes and new lines may be added at will. Don’t forget, however, to place the symbol # in every line corresponding to a comment.
- The script is divided into a few parts. These parts have capital letter titles within brackets. Don’t touch them!
- Don’t remove any line that is not a comment. See below how to appropriately edit these command lines.
Let’s take a look now at each part of a script (this is the script .epg demo file):
Warning
All variables defined in a .epg are case-insensitive. Consider this fact when naming your model’s variables.
Part 1: THE WORLD¶
The first section of the script is titled: THE WORLD. An example of its content is shown:
shapefile = ['riozonas_LatLong.shp','nome_zonas','zona_trafe']
edges = edges.csv
sites = sites.csv
encoding =
where,
- shapefile
- Is a list with 3 elements: the first is the path, relative to the working directory, of the shapefile file;
the second is the variable, in the shapefile, which contains the names of the localities (polygons of the map); the third and last is the variable, in the shapefile, which contains the geocode of the localities. If you don’t have a map for you simulation, leave the list empty: location = [] ). Epigrass also accepts Maps in the Geopackage format. Epigrass assume that maps use the EPSG:4326 projection. Using other projection may result in errors related to the visualizaton of results. edges
This is the name of the .CSV (comma-separated-values) file containing the list of edges and their attributes.
- sites
- This is the name of the .CSV file containing the list of sites and their attributes.
- encoding
- This is the encoding used in your sites and edges files. This is very important when you use location names
which include non-ascii characters. The default encoding is latin-1. If you use any other encoding, please specify it here. Example: utf-8.
Note
All paths in the .epg file are relative to the working directory.
Part 2: EPIDEMIOLOGICAL MODEL¶
This is the main part of the script. It defines the epidemiological model to be run. The script reads:
modtype = SIR
Here, the type of epidemiological model is defined, in this case is a deterministic SIR model. Epigrass has some built-in models:
| Name | Determ. | Stochastic |
|---|---|---|
| Susceptible-Infected-Recovered | SIR | SIR_s |
| Susceptible-Exposed-Infected-Recovered | SEIR | SEIR_s |
| Susceptible-Infected-Susceptible | SIS | SIS_s |
| Susceptible-Exposed-Infected-Susceptible | SEIS | SEIS_s |
| SIR with fraction with full immunity | SIpRpS | SIpRpS_s |
| SEIR with fraction with full immunity | SEIpRpS | SEIpRpS_s |
| SIR with partial immunity for all | SIpR | SIpR_s |
| SEIR with partial immunity for all | SEIpR | SEIpR_s |
| SIR with immunity wane | SIRS | SIRS_s |
A description of these models can be found in the chapter Epidemiological modeling. The stochastic versions of models use Poisson distribution as default for the number of new cases (L(t+1)), They are not full stochastic models. Besides these, the user may define his/her own model and access it by setting modtype = Custom.
Part 3: MODEL PARAMETERS¶
The epidemic model is defined by variables and parameter which require initialization:
#==============================================================#
# They can be specified as constants or as functions of global or
# site-specific variables. Site-specific variables are provided
# in the sites .csv file. In this file, all columns after the 4th
# are collected into a values tuple, which can be referenced here
# as values[0], values[1], values[2], etc.
# Examples:
# beta = 0.001
# beta=values[0] #assigns the first element of values to beta
# beta=0.001*values[1]
# beta=0.001*N # N is a global name for total site population
# Currently, Epigrass requires that parameters beta, alpha, e, r, delta, B, w, p
# be present in the .epg even if they will not be used. Do not erase these lines.
# Just disregard them if they are not useful to you.
beta = 0.4 #transmission coefficient (contact rate * transmissibility)
alpha = 1 # clumping parameter
e = 1 # inverse of incubation period
r = 0.1 # inverse of infectious period
delta = 1 # probability of acquiring full immunity [0,1]
B = 0 # Birth rate
w = 0 # probability of immunity waning [0,1]
p = 0 #
These are the model parameters. Not all parameters are necessary for all models. For example, e is only required for SEIR-like models. Don’t remove the line, however because that will cause an error. We recommend that, if the parameter is not necessary, just add a comment after it as a reminder that it is not being used by the model.
In some cases, one may wish to assign site-specific parameters. For example, transmission rate may be different between localities that are very distant and are exposed to different climate. In this case site specific variables can be added as new columns to the site file. All columns after the geocode are packed into a tuple named values and can be referenced in the order they appear. I.e., the first element of the tuple is values[0], the second element is values[1], the third element is values[2] and so on.
Part 4: INITIAL CONDITIONS
In this part of the script, the initial conditions are defined. Here, the number of individuals in each epidemiological state, at the start of the simulation, is specified. It reads:
#==============================================================#
# Here, the number of individuals in each epidemiological
# state (SEI) is specified. They can be specified in absolute
# or relative numbers.
# N is the population size of each site.
# The rule defined here will be applied equally to all sites.
# For site-specific definitions, use EVENTS (below)
# Examples:
# S,E,I = 0.8*N, 10, 0.5*N
# S,E,I = 0.5*N, 0.01*N, 0.05*N
# S,E,I = N-1, 1, 0
S = N
E = 0
I = 0
Here, N is the total population in a site (as in the datafile for sites). In this example, we set all localities to the same initial conditions (all individuals susceptible) and use an event (see below) to introduce an infectious individual in a locality. The number of recovered individuals is implicit, as R = N-(S+E+I)
Another possibility is to define initial conditions that are different for each site. For this, the data must be available as extra columns in the site data file and these columns are referenced to using the values tuple explained above.
Part 5: EPIDEMIC EVENTS¶
The next step is to define events that will occur during the simulation. These events may be epidemiological (arrival of an infected, for example) or a public health action (vaccination campaign, for example):
#=============================================================#
# Specify isolated events.
# Localities where the events are to take place should be Identified by the geocode, which
# comes after population size on the sites data file.
# All coverages must be a number between 0 and 1.
# Seed : [('locality1's geocode',epid state, n),('locality2's geocode', epid state, n),...]
# N infected cases will be added to locality at time 0.
# Vaccinate: [('locality1's geocode', [t1,t2,...], [cov1,cov2,...]),('locality2's geocode', [t1,t2,...], [cov1,cov2,...])]
# Quarantine: [(locality1's geocode,time,coverage), (locality2's geocode,time,coverage)]
#seed = [(4550601,'ip20',10)] #santo cristo #
seed = [(4552110,'ip20',10)] #pechincha #
#seed = []
Vaccinate = [] #
Quarantine = []
The events currently implemented are:
- seed
- One or more infected individual(s) are introduced into a site, at the beggining of the simulation. The notation for a single event is: Seed = [(‘locality1’s geocode’,epid state, n),(‘locality2’s geocode’, epid state, n),…]. For example, seed = [(2,’I’,10)] programs the arrival of 10 infected individuals at site geocode 2, at time 1.
- Vaccinate
- Implements a campaign that vaccinates a fraction of the population in a site, at a pre-defined time-step. For multiple events, the notation is: [(‘locality1’s geocode’, [t1,t2,…], [cov1,cov2,…]),(‘locality2’s geocode’, [t1,t2,…], [cov1,cov2,…])], where the first element of every triplet is the geocode of the city, the second element is a list of the time(s) when the campaign is carried on, and the third element is the coverage(s). For example, the event [(2,[10],[0.7])] means that city 2, at time 10, has 70% of its population vaccinated. Mathematically, it means (in the model), the removal of individuals from the susceptible to the recovered stage.
- Quarantine
- Prevents any individual from leaving a site, starting at t. Currently disabled.
Part 6: TRANSPORTATION MODEL¶
Here, there are two options regarding the movement of infected individuals from site to site (through the edges). If stochastic = 0, the process is simulated deterministically. The number of infected passengers commuting through an edge is a fraction p of the infected population that is traveling. p is calculated as total passengers/total population.
If stochastic = 1, the number of passengers is sampled from a Poisson distribution with parameter given by the expected number of travelling infectives (calculated as above):
#=========================================================#
# If doTransp = 1 the transportation dynamics will be
# included. Use 0 here only for debugging purposes.
doTransp = 1
# Mechanism can be stochatic (1) or deterministic(0).
stochastic = 1
#Average speed of transportation system in km per time step. Enter 0 for instantaneous travel.
#Distance unit must be the same specified in edges files
speed =0 #1440 km/day -- equivalent to 60 km/h
That ends the definition of the model.
Part 7: SIMULATION AND OUTPUT¶
Now it is time to define some final operational variables for the simulation:
#==============================================================#
# Number of steps
steps = 50
# Output dir. Must be a full path. If empty the output will be generated on the
# same path as the model script.
outdir =
# Output file
outfile = simul.dat
# Database Output
# SQLout can be 0 (no database output) or 1
SQLout = 1
# Report Generation
# The variable report can take the following values:
# 0 - No report is generated.
# 1 - A network analysis report is generated in PDF Format.
# 2 - An epidemiological report is generated in PDF Format.
# 3 - A full report is generated in PDF Format.
# siteRep is a list with site geocodes. For each site in this list, a detailed report is apended to the main report.
report = 0
siteRep = []
# Replicate runs
# If replicas is set to an integer(n) larger than zero, the model will be run
# n times and the results will be consolidated before storing.
# RandSeed = 1: the seed will be randomized on each replicate
# RandSeed = 2: seeds are taken sequentially from the site's file
# Note: Replicate mode automatically turn off report and batch options.
Replicas = 10
RandSeed = 2
#Batch Run
# list other scripts to be run in after this one. don't forget the extension .epg
# model scripts must be in the same directory as this file or provide full path.
# Example: Batch = ['model2.epg','model3.epg','/home/jose/model4.epg']
Batch = []#['sarsDF.epg','sarsPA.epg','sarsRS.epg']
where,
- step
- Number of time steps for the simulation.
- outdir
- Directory for data output (currently not in use)
- outfile
- .csv filename that can be imported into R as a dataframe. This .csv file contains the simulated timeseries for all nodes.
- sqlout
- Use sqlout = 1 if simulated time series are to be stored. Time series of L, S, E, and I, from simulations,
are stored in a SQLite database named Epigrass.sqlite. The results of each individual simulation is stored in a different
table named after the model’s script name, the date and time the simulation has been run. For instance, suppose you run
a simulation of a model stored in a file named script.epg, then at the end of the simulation, a new table in the
epigrass database will be created with the following name: script_Wed_Jan_26_154411_2005. Thus, the results of
multiple runs from the same model get stored independently.
Batch=[]
Script files included in this list are executed after the currently file is finished.
Operation¶
For running simulations, after all the information has been entered and checked on the first tab of the GUI, you can press the Run button to start the simulation or the :guilabel`Exit` button. When the Run button is pressed, the ~/.epigrassrc file is updated with all the information entered in the GUI. If the Exit button is pressed, all information entered since the last time the Run button was pressed is lost.
Epigrass also allows running simulation straight from the command line, with the epirunner executable. all you have to do is:
$ epirunner mymodel.epg
and the model will executed with the settings specified in the ~/.epigrassrc file. for help with epirunner, type:
$ epirunner -h
No Custom Model Available.
usage: epirunner [options] your_model.epg
Run epigrass models from the console
positional arguments:
EPG Epigrass model definition file (.epg).
optional arguments:
-h, --help show this help message and exit
-b <mysql|sqlite|csv>, --backend <mysql|sqlite|csv>
Define which datastorage backend to use
-u DBUSER, --dbusername DBUSER
MySQL user name
-p DBPASS, --password DBPASS
MySQL password for user
-H DBHOST, --dbhost DBHOST
MySQL hostname or IP address
--upload UPLOAD Upload your models and latest simulation to Epigrass Web
-P, --parallel use multiprocessing to run the simulation
-D, --dashboard Open dashboard on browser after th run is done.
-V, --view-only Only Open dashboard.
The Web Dashboard¶
To interact and explore the simulations stored in the database, Epigrass offers an interactive dashboard. It allows you to navigate to any simulation previously done for a given model, and inspect it interactively. You can ask for it to open right after a simulation is done:
$ epirunner -D mymodel.epg
or you can open it for a given model by typing from the directory where the epg file is located:
$ epirunner -V mymodel.epg
The Dashboard shows various aspects of the Model output, such as Choroplectic map with the final state of the simulation,
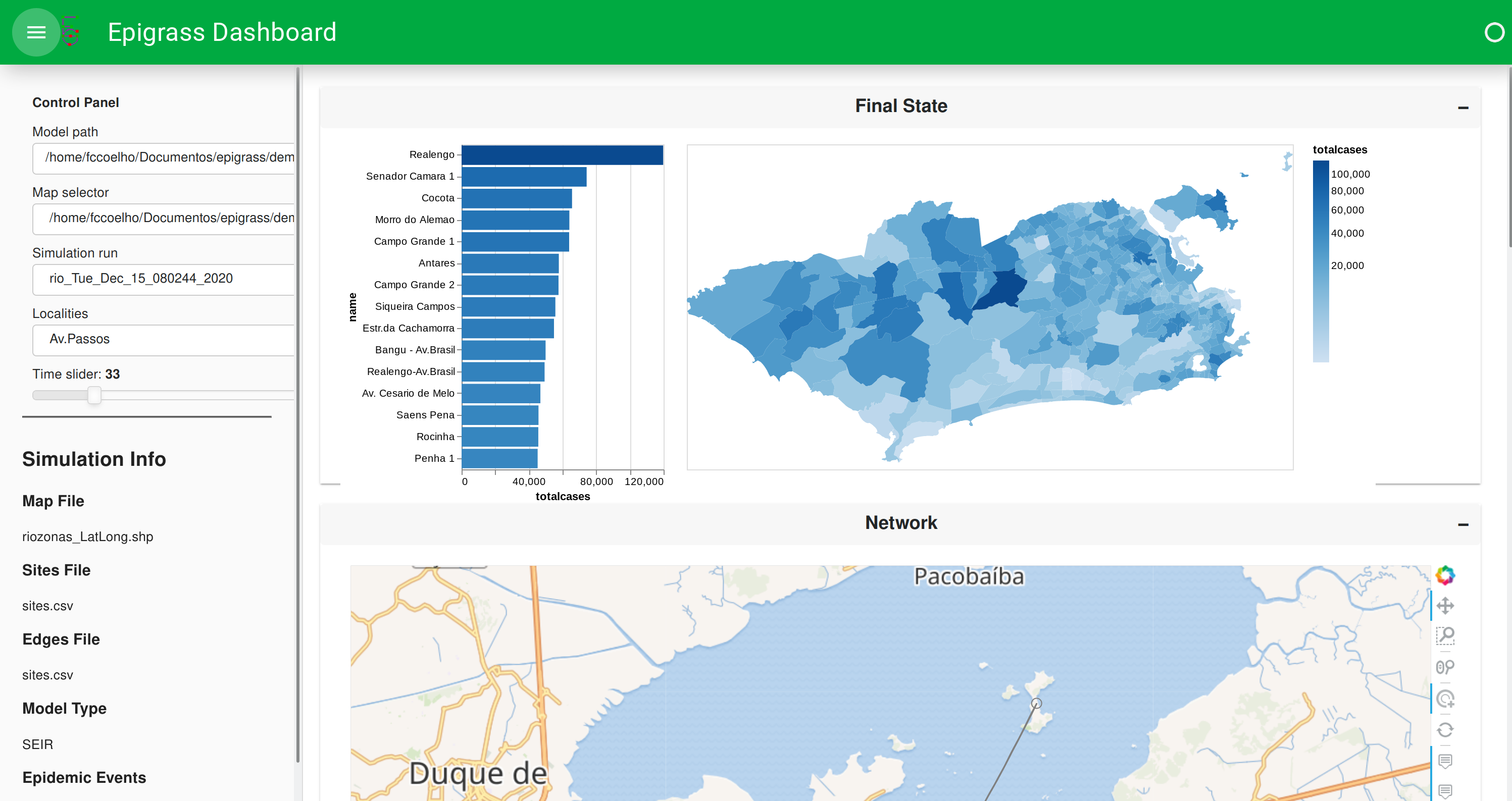
Figure: Dashboard showing map with the final state of the model.
It also shows the network neighborhood of any of the localities of the models
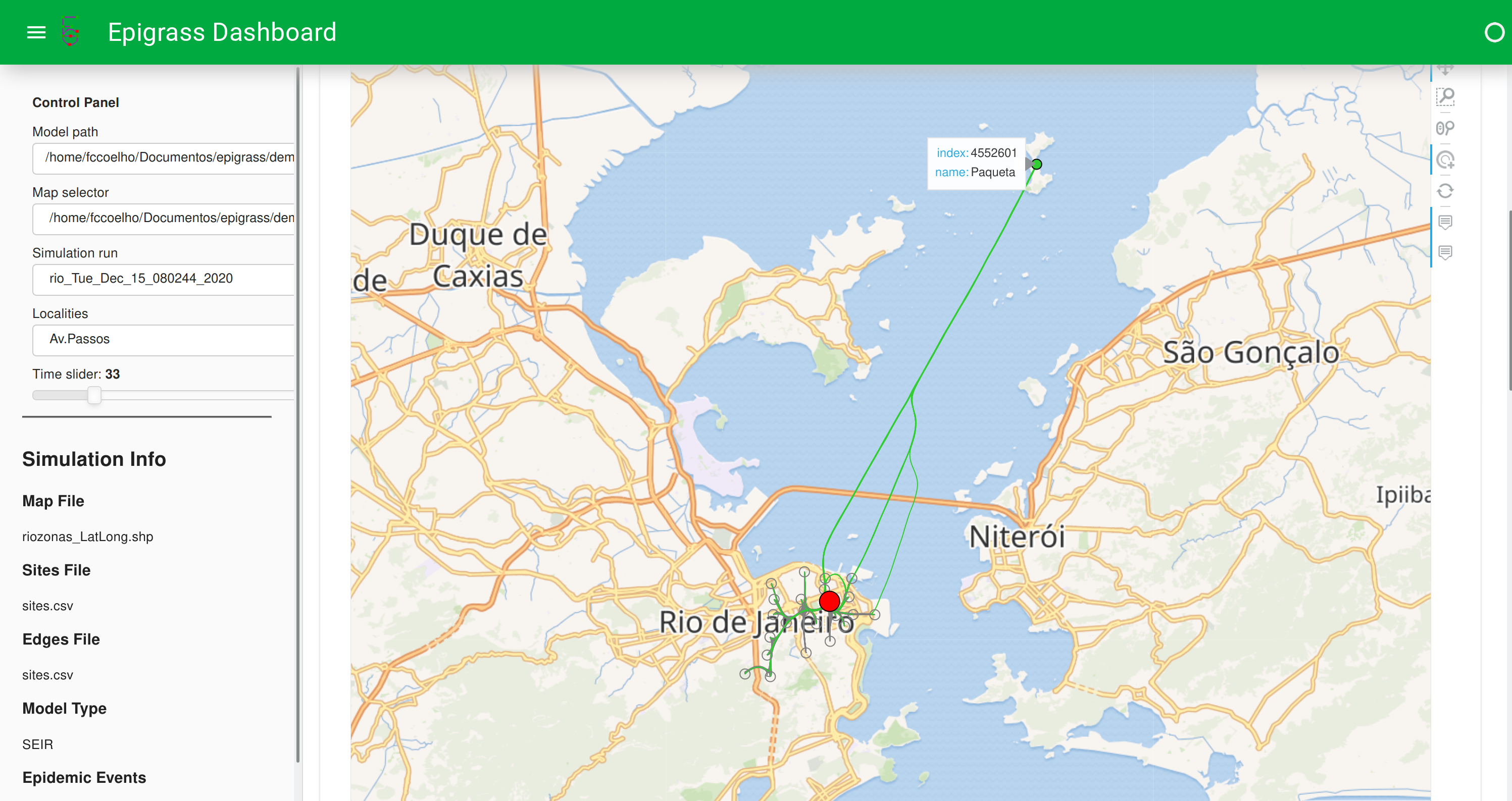
Figure: Dashboard showing network neighborhood of a locality marked in red.
It also shows a map which can be animated and timeseries plots of all the state variables
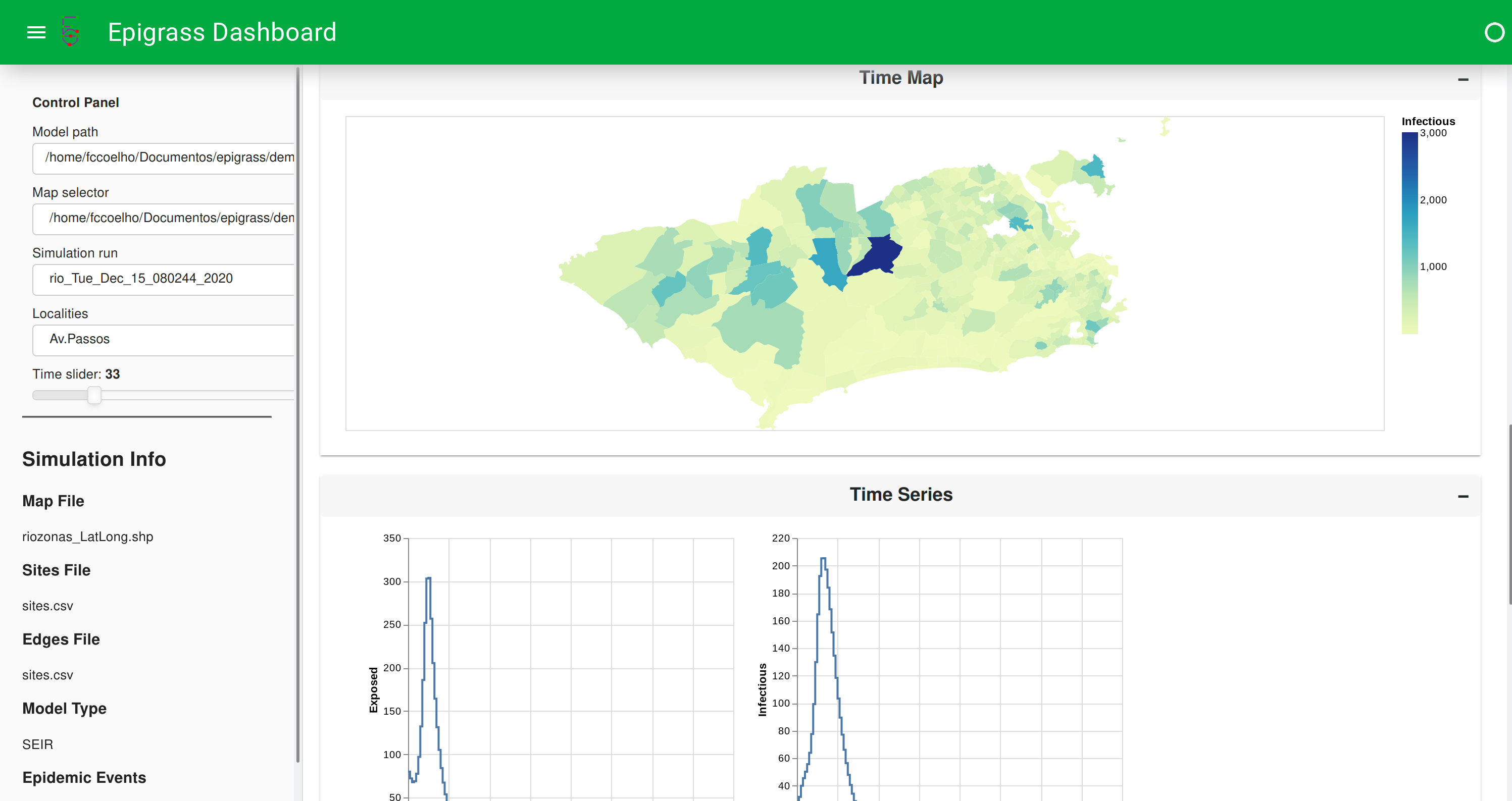
Figure: Dashboard showing timeseries visualizations.